The 4 ways to run Jupyter Notebook in command line
Jupyter Notebook is a perfect tool for data analysis and quick prototyping. You can easily mix Python (or R, Julia, Scala ...) code with Markdown. What to do if you would like to run your .ipynb file in the command line? There are several approaches to execute notebook in the terminal. There is no one-size fits all, it all depends on your needs. Do you need to execute notebook and share the output as PDF or HTML file? Would you like to upload executed notebook to the cloud storage? Would you like to add parameters to your notebook? I hope you will find solution for yourself in our list.
1. Execute notebook with nbconvert
The nbconvert is an open-source tool for converting and executing Jupyter Notebooks. It is already installed with Jupyter Notebook (it is used internally to convert .ipynb files to other formats in the web User Interface). The nbconvert provides Python API and command line tool. Just run the following command to see help message:
jupyter nbconvert
If you would like to execute the notebook and overwrite your existing notebook with most recent computations then please use the following command:
jupyter nbconvert --execute --to notebook --inplace your-notebook.ipynb
It will execute your-notebook.ipynb and overwrite it with new values.
Below is the example notebook that I'm using in my examples:
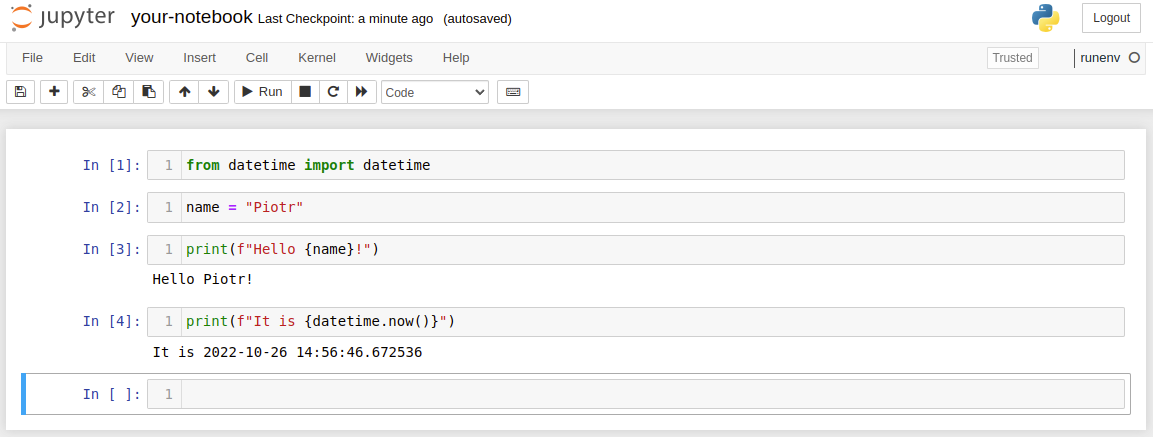
If you would like to execute the your-notebook.ipynb and save most recent version in other .ipynb file please use the command:
jupyter nbconvert --execute --to notebook your-notebook.ipynb >> [NbConvertApp] Converting notebook your-notebook.ipynb to notebook >> [NbConvertApp] Writing 2538 bytes to your-notebook.nbconvert.ipynb
It will save executed notebook to your-notebook.nbconvert.ipynb file. You can specify the custom output name and custom output directory:
jupyter nbconvert --execute --to notebook --output custom-name --output-dir /custom/path/ your-notebook.ipynb
The resulting notebook will be save to the custom-name.ipynb file at /custom/path/. Please notice that in the command line there is no file extension in the --output parameter (just custom-name, not custom-name.ipynb).
The next useful flag is --allow-errors. With this flag, all cells will be executed, even if one of them will have error. The default behavior is to stop execution after first error and don't create output notebook. The example command that execute notebook even with errors:
jupyter nbconvert --execute --to notebook --allow-errors your-notebook.ipynb
What if you would like to share executed notebook with others? You can execute the notebook and save output into PDF or HTML format. Additionally, you can hide code in the final notebook. The example command that will execute notebook and save it as HTML file with code hidden.
jupyter nbconvert --execute --to html --no-input your-notebook.ipynb >> [NbConvertApp] Converting notebook your-notebook.ipynb to html >> [NbConvertApp] Writing 576249 bytes to your-notebook.html
The your-notebook.html file is created, you can open it in your web browser by double click.
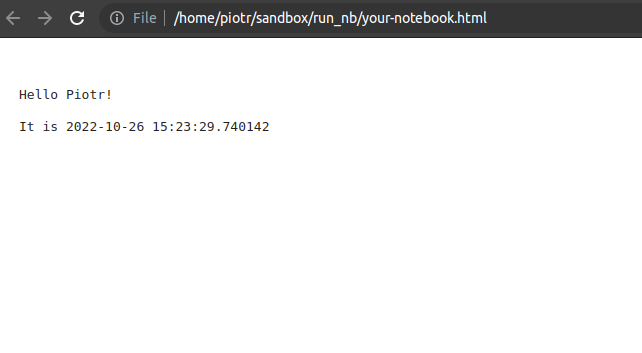
Executing and exporting to the PDF might be more tricky (other ways to export notebook to PDF) because you need to install additional packages. The nbconvert can export notebook to PDF in two ways:
- by exporting to LaTeX and then to PDF,
- be exporting to HTML and then to PDF.
I prefer the latter method because requires less packages to be installed.
You need to install pyppeteer package with following command:
pip install nbconvert[webpdf]
The command line to execute notebook and save output to the PDF file without input code:
jupyter nbconvert --execute --to webpdf --no-input your-notebook.ipynb >> [NbConvertApp] Converting notebook your-notebook.ipynb to webpdf >> [NbConvertApp] Building PDF >> [NbConvertApp] PDF successfully created >> [NbConvertApp] Writing 17243 bytes to your-notebook.pdf
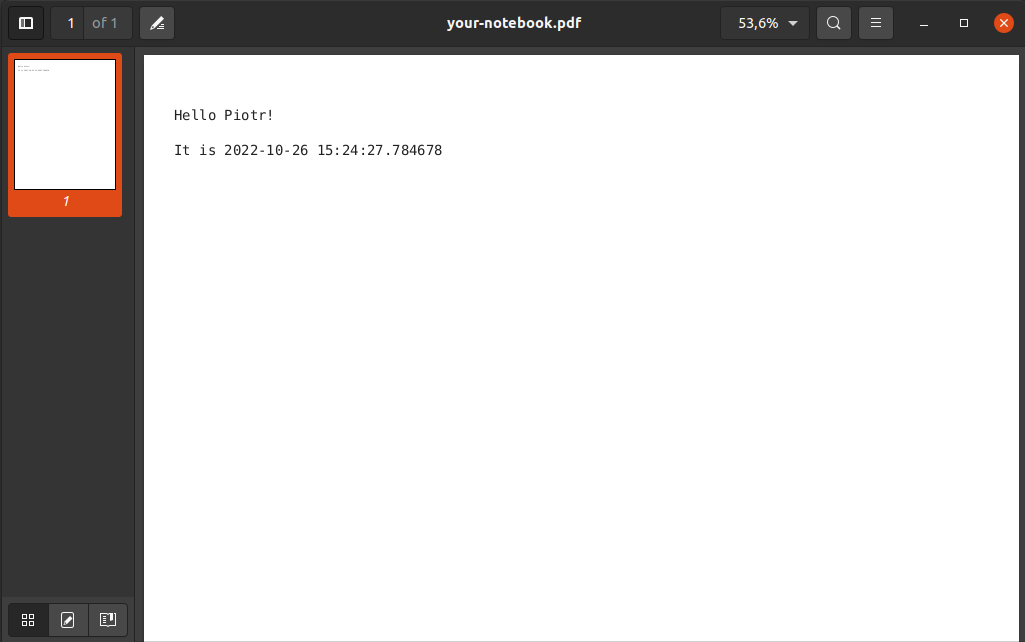
The nbconvert tool is really amazing.
2. Execute parameterized notebook with papermill
The papermill tool allows you to execute parameterized notebooks. You need to install papermill as additional package:
pip install papermill
You need to set parameters cell in the notebook to be able to execute parameterized notebook in the command line. Please click View->Cell Toolbar->Tags:
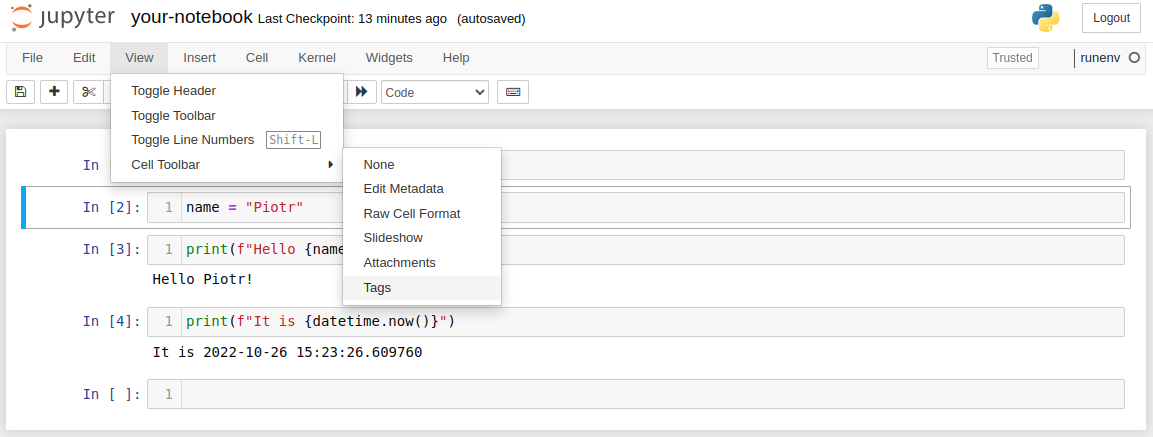
Please add parameters tag:
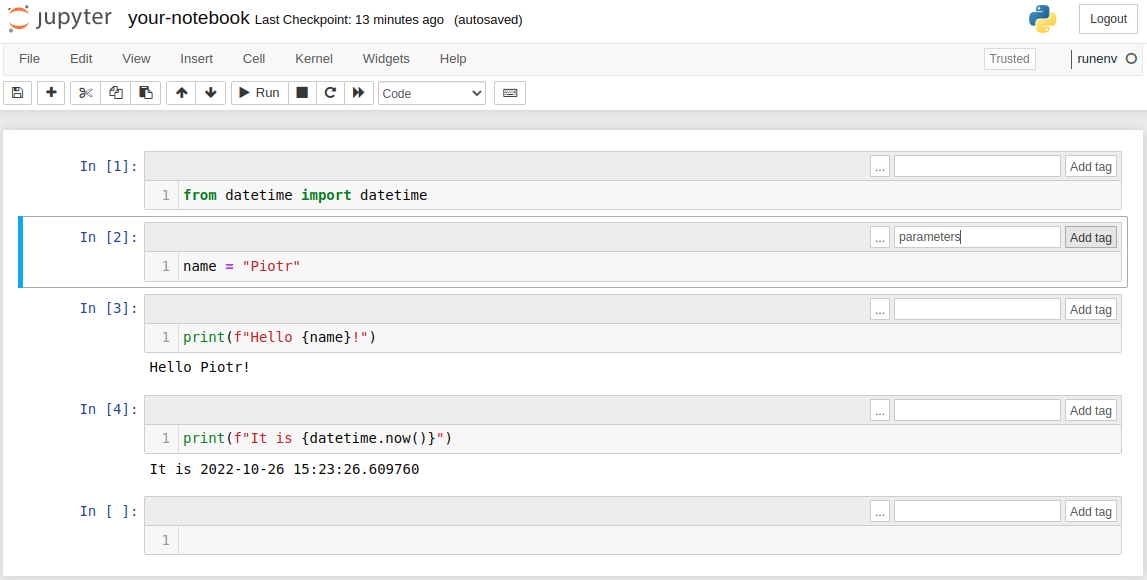
The notebook after inserting tag:
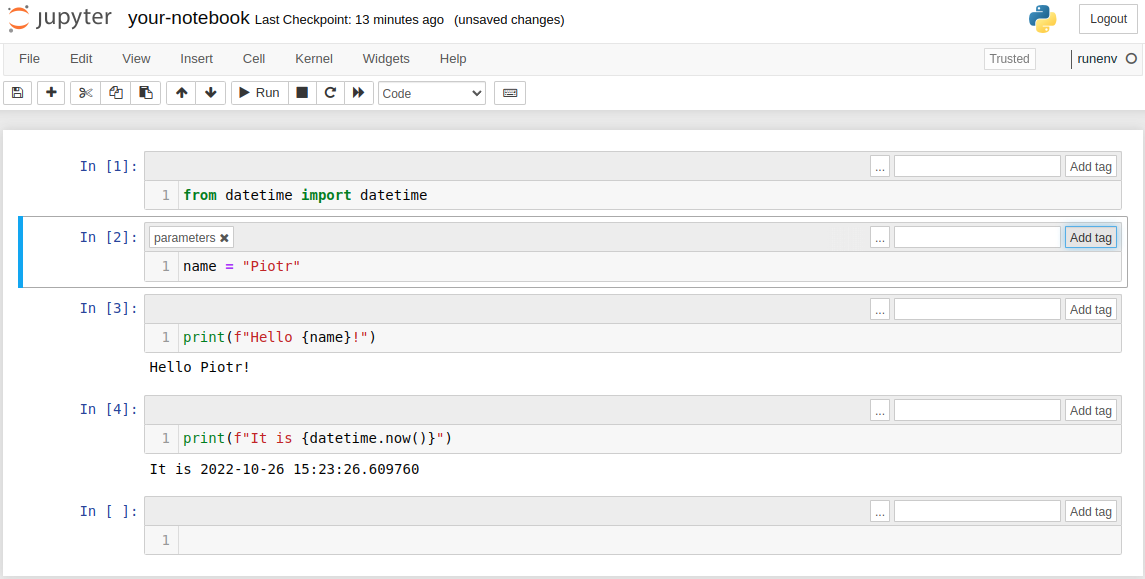
You can execute the notebook with parameters in the command line. The name of the parameter is the same as variable:
papermill -p name Piotrek your-notebook.ipynb output-notebook.ipynb >> Input Notebook: your-notebook.ipynb >> Output Notebook: output-notebook.ipynb >> Black is not installed, parameters wont be formatted >> Executing: 0%| | 0/6 [00:00<?, ?cell/s]>> Executing notebook with kernel: runenv >> Executing: 100%|█████████████████████████████████████████████████████████████████████████████████████████████████████| 6/6 [00:01<00:00, 5.35cell/s]
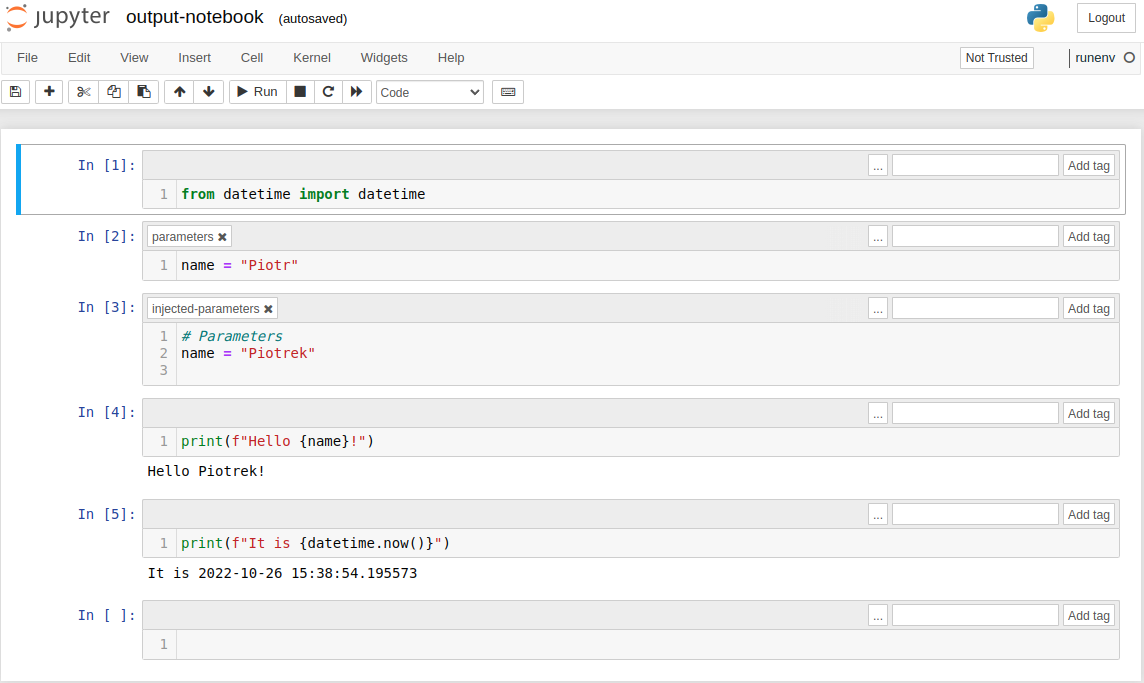
The final notebook is saved to output-notebook.ipynb. Please notice that output notebook has one more cell. It has parameters as variables and injected-parameters tag:
If you would like to execute the notebook with parameters and then export to PDF or HTML or other formats then you can build a pipeline with nbconvert:
- step 1: run parameterized notebook with
papermill, - step 2: convert output notebook to desired format with
nbconvert.
The papermill offers you option to store the output file in the cloud storage, please check documentation for details.
3. Manually convert notebook to .py script
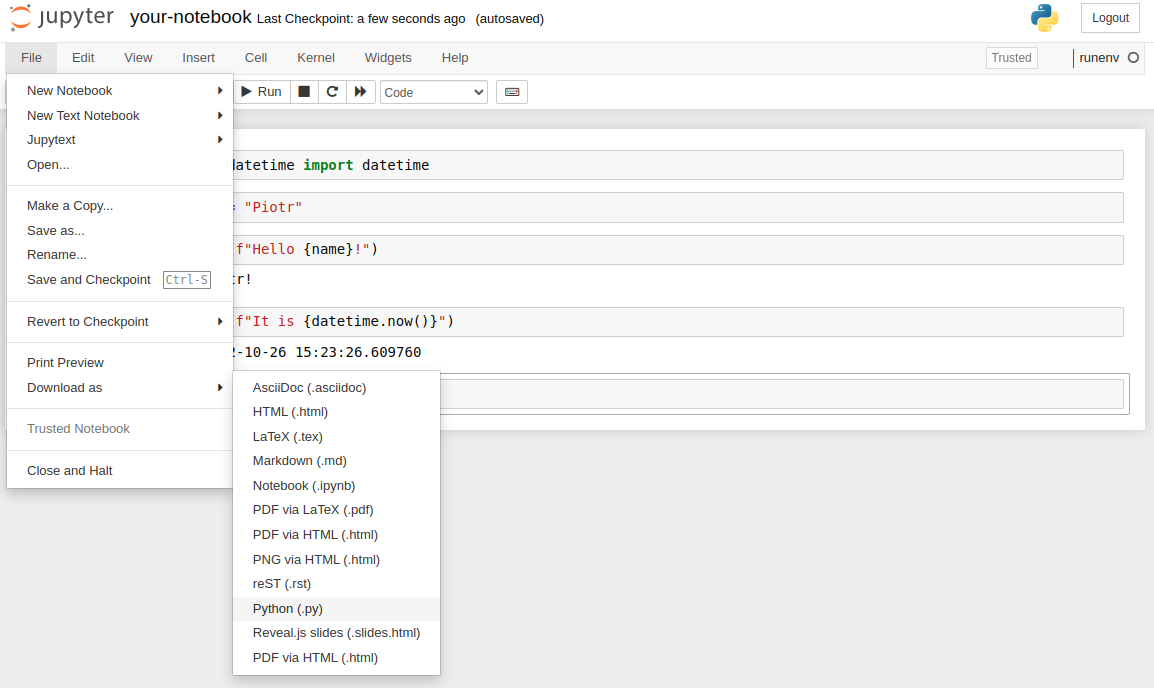
The next approach is to manually convert Jupyter Notebook to Python .py script. Please click File->Download as->Python (.py):
You will download the .py script automatically created based on your notebook (it is done internally with nbconvert). Below is the example .py script:
#!/usr/bin/env python # coding: utf-8 # In[1]: from datetime import datetime # In[2]: name = "Piotr" # In[3]: print(f"Hello {name}!") # In[4]: print(f"It is {datetime.now()}") # In[ ]:
Please notice that each cell is marked as # In [<cell number>] in comments. You can add execute permission to the file and run it as a script:
chmod +x your-notebook.py ./your-notebook.py >> Hello Piotr! >> It is 2022-10-26 16:05:30.735248
4. jupytext pair script with notebook
The manual download of a script from the notebook might be tedious, especially after notebook updates. There is a tool called jupytext that allows you to keep synchronized .py script with .ipynb notebook. It can be easily installed:
pip install jupytext
Please enable synchronization after installation by clicking File->Jupytext->Pair Notebook with light script:
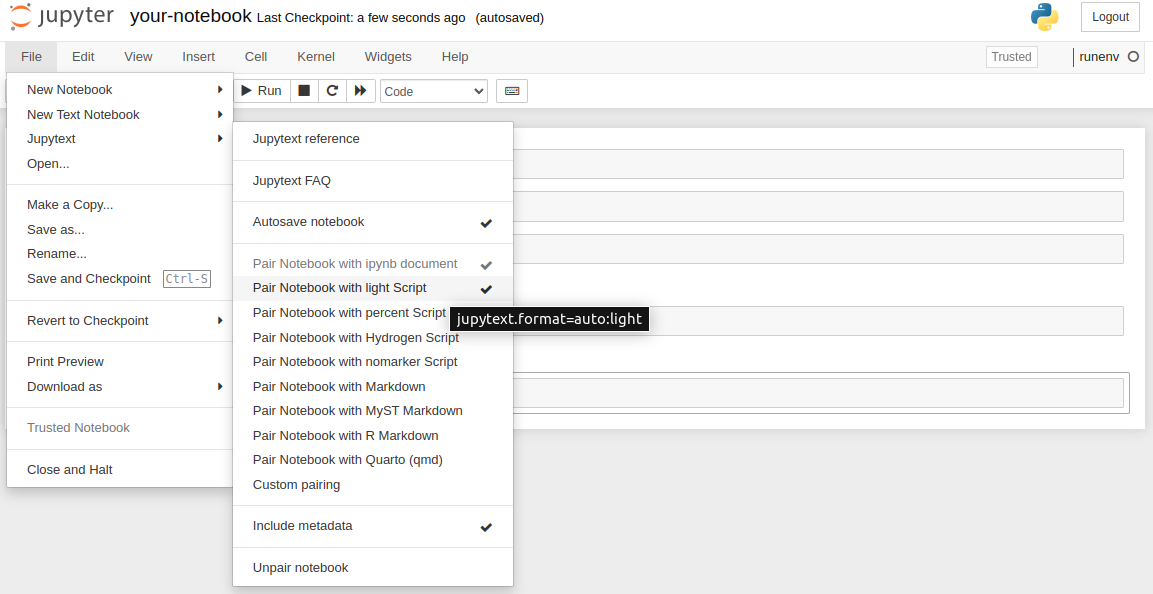
There will be created your-notebook.py file that is updated after each notebook save. The example .py script file created with jupytext.
# --- # jupyter: # jupytext: # formats: ipynb,py:light # text_representation: # extension: .py # format_name: light # format_version: '1.5' # jupytext_version: 1.14.1 # kernelspec: # display_name: runenv # language: python # name: runenv # --- from datetime import datetime name = "Piotr" print(f"Hello {name}!") print(f"It is {datetime.now()}")
You can read more about available synchronization options in the jupytext documentation.
Summary
There are many ways in which you can execute Jupyter Notebook in terminal. The nbconvert tool provides many options for exporting notebook to different formats, like PDF or HTML. It can be used to hide code in out notebook. The papermill framework allows to easily parameterize notebooks. You can convert notebook to .py file and execute it as plain Python script. It can be done manually or with jupytext package. I hope you will find your way for running Jupyter Notebooks in the command line.
You might be interested in the open-source framework for converting Jupyter Notebooks to web applications that we are working on. It is called Mercury. You can use it to share your notebooks with non-technical users. You can read more about Mercury on our website.
Run AI for Machine Learning — Fully Local
MLJAR Studio is a private AI Python notebook for data analysis and machine learning. Generate code with AI, run experiments locally, and keep full control over your workflow.
About the Authors


Related Articles
- Schedule Jupyter Notebook to run daily
- Create Presentation from Jupyter Notebook
- Create Parameterized Presentation in Jupyter Notebook
- The 3 ways to hide code in Jupyter Notebook Presentation
- Convert Jupyter Notebook to spaCy NLP web application
- The 2 ways to convert Jupyter Notebook Presentation to PDF slides
- The 3 ways to share password protected Jupyter Notebook
- The 3 ways to change figure size in Matplotlib
- The 5 ways to publish Jupyter Notebook Presentation
- The 2 ways to save and load scikit-learn model