4 Effective Ways to Visualize Random Forest

A Random Forest is a supervised algorithm used in machine learning. It operates by constructing a multitude of decision trees (typically using a binary tree structure where each node has two children) and combining their results to improve accuracy and prevent overfitting. In a Random Forest, each data sample is passed through multiple decision trees, each of which assigns a target value to the sample. The final prediction is typically determined by aggregating the outputs of all the individual trees, such as taking the majority vote for classification or averaging the predictions for regression.
The trees in the forest make decisions based on the features of the input sample. Each tree is built independently using a random subset of the data and a random selection of features, which helps to create diversity and reduce correlation between the trees. This process helps to make Random Forest more robust than individual decision trees.
The Random Forest algorithm can be divided into two types based on the target values:
- Classification forests used to classify samples into a set of distinct classes. In scikit-learn, this is handled by the
RandomForestClassifier. - Regression forests used to predict continuous numerical values within a range. In scikit-learn, this is handled by the
RandomForestRegressor.
Random Forests are widely used due to their ability to handle large datasets, manage feature importance, and provide high accuracy while reducing overfitting compared to a single decision tree. They are also valuable for decision-making processes because of their ensemble approach and visual interpretability through feature importance and tree visualization.
Below I show how to use scikit-learn and 4 ways to plot a tree from it:
plot_tree- req. matplotlibgraphviz- req. graphvizdtreeviz- req. dtreeviz and graphvizsupertree- req. supertree
Classification task
Prepare data
In this example let's use a real classic - iris dataset.
from sklearn.ensemble import RandomForestClassifier from sklearn.datasets import load_iris from sklearn.model_selection import train_test_split from sklearn.metrics import accuracy_score # Load the Iris dataset iris = load_iris() # Separate features (X) and target labels (y) X = iris.data # Features (flower measurements) y = iris.target # Labels (species) # Define feature names and target class for clarity features = iris.feature_names # List of feature names (petal length, etc.) target = 'species' # Target variable (species of the flower) class_names = iris.target_names # List of species (setosa, versicolor, virginica) # Split the dataset into training and testing sets # test_size=0.2 means 20% of the data will be used for testing # random_state ensures reproducibility X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
Train model
# Initialize the RandomForestClassifier rf_classifier = RandomForestClassifier( n_estimators=50, # Number of trees in the forest max_depth=5, # Maximum depth of the trees random_state=42 # Seed for reproducibility ) # Train the classifier on the training data rf_classifier.fit(X_train, y_train)
Plot with plot_tree
First method I use comes from sklearn package and to work matplotlib is required. It allows us to easily produce figure of the tree (without intermediate exporting to graphviz).
import matplotlib.pyplot as plt from sklearn.tree import plot_tree # Plot the tree using the plot_tree function from sklearn tree = rf_classifier.estimators_[0] plt.figure(figsize=(20,10)) # Set figure size to make the tree more readable plot_tree(tree, feature_names=features, # Use the feature names from the dataset class_names=class_names, # Use class names (species names) filled=True, # Fill nodes with colors for better visualization rounded=True) # Rounded edges for nodes plt.title("Decision Tree from the Random Forest") plt.show()
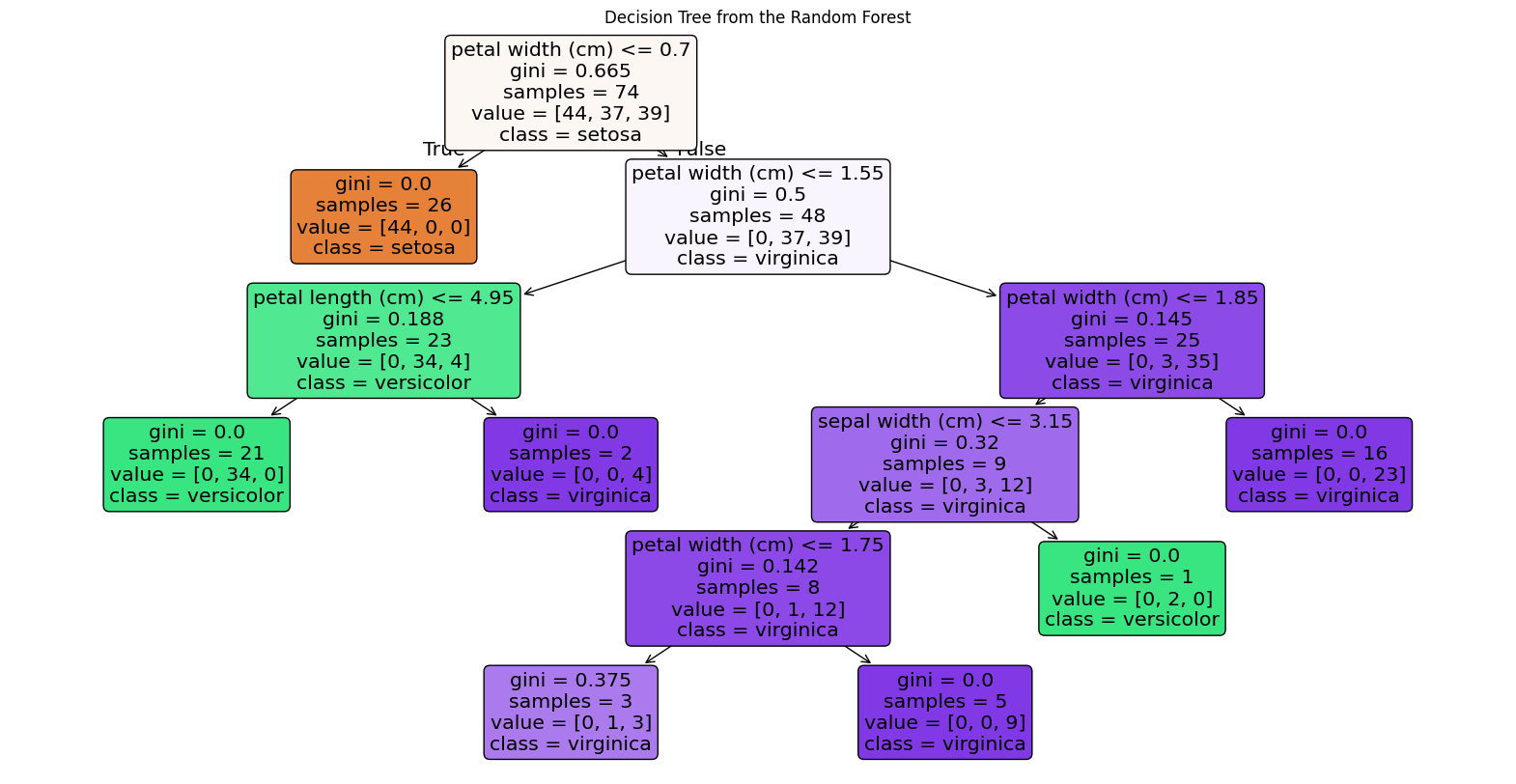
Plot with graphviz
graphviz outpur may look simmilar to plot_tree() but is more readable. Additional work that needs to be done is exporting my tree to a DOT file.
from sklearn.tree import export_graphviz import graphviz # Assuming rf_classifier is already trained # Select one tree from the random forest (e.g., the first one) tree = rf_classifier.estimators_[0] # Export the selected tree to DOT format dot_file_path = "random_forest_tree.dot" export_graphviz( tree, out_file=dot_file_path, feature_names=features, # Feature names from the Iris dataset class_names=class_names, # Class names (setosa, versicolor, virginica) filled=True, # Color the nodes rounded=True, # Rounded corners for nodes special_characters=True # Allows special characters in labels ) with open(dot_file_path) as f: dot_graph = f.read() # Visualize the tree using graphviz graph = graphviz.Source(dot_graph) graph.render("random_forest_tree") graph # Display the tree graph
Plot with dtreeviz
To use dtreeviz don't forget to install graphviz. With it I not only skip converting tree, but also it creates diagrams within plot. Plots made with dtreeviz are elegant and insightful.
from dtreeviz import model tree = rf_classifier.estimators_[0] viz_model = model( tree, X_train=X_train, y_train=y_train, target_name=target, class_names=list(class_names), tree_index=2 ) viz_model.view()
Plot with supertree
supertree is single INTERACTIVE package on my list of ways to visualize tree from random forests.
It's easy to use and in my humnle opinion is the most visually appealing out of all 4 packages.
from supertree import SuperTree st = SuperTree( rf_classifier, X_train, y_train, iris.feature_names, iris.target_names ) st.show_tree(which_tree=0)
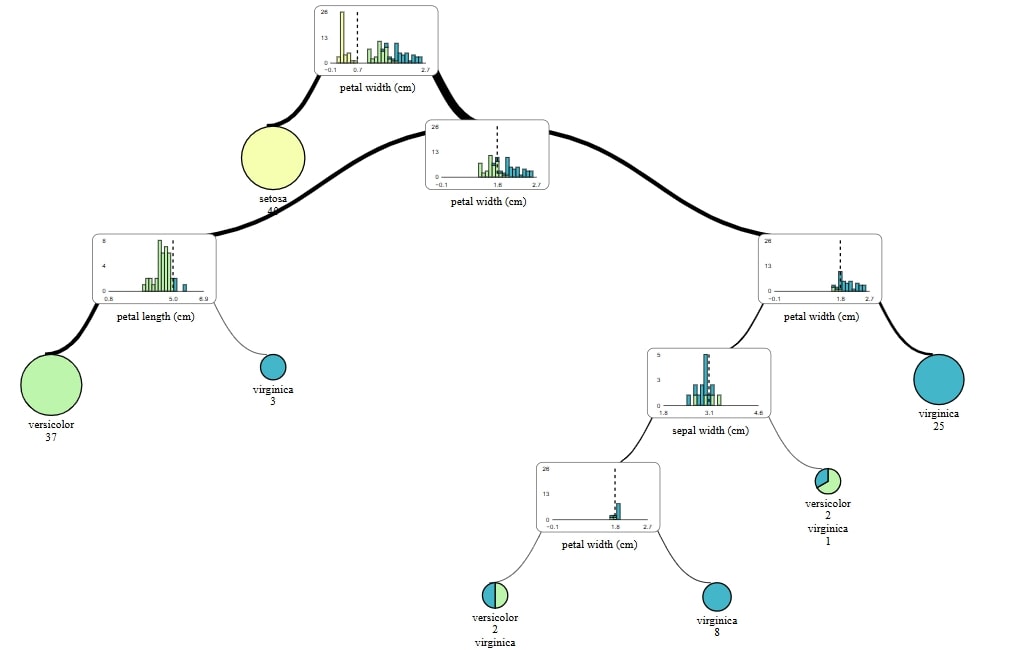
Regression
For regression task I'm going to use diabetes data.
Becuase this examples are very simmilar, I will limit myself to comments in code.
Train model
from sklearn.ensemble import RandomForestRegressor from sklearn.datasets import load_diabetes from sklearn.model_selection import train_test_split from sklearn.tree import export_graphviz import graphviz # Load the diabetes dataset diabetes = load_diabetes() # Separate features (X) and target (y) X = diabetes.data # Features (e.g., age, BMI, etc.) y = diabetes.target # Target (a continuous value, which is a measure of disease progression) # Split the dataset into training and testing sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42) # Initialize the RandomForestRegressor rf_regressor = RandomForestRegressor( n_estimators=50, # Number of trees in the forest max_depth=5, # Maximum depth of the trees random_state=42 # Seed for reproducibility ) # Train the regressor on the training data rf_regressor.fit(X_train, y_train)
Plot with plot_tree
import matplotlib.pyplot as plt from sklearn.tree import plot_tree # Assuming rf_regressor is a trained RandomForestRegressor tree = rf_regressor.estimators_[0] # Load the diabetes dataset diabetes = load_diabetes() # Set figure size plt.figure(figsize=(20, 10)) # Plot the tree plot_tree(tree, feature_names=diabetes.feature_names, # Use the feature names from the dataset filled=True, # Fill nodes with colors for better visualization rounded=True) # Rounded edges for nodes # Add a title plt.title("Decision Tree from the Random Forest") plt.show()
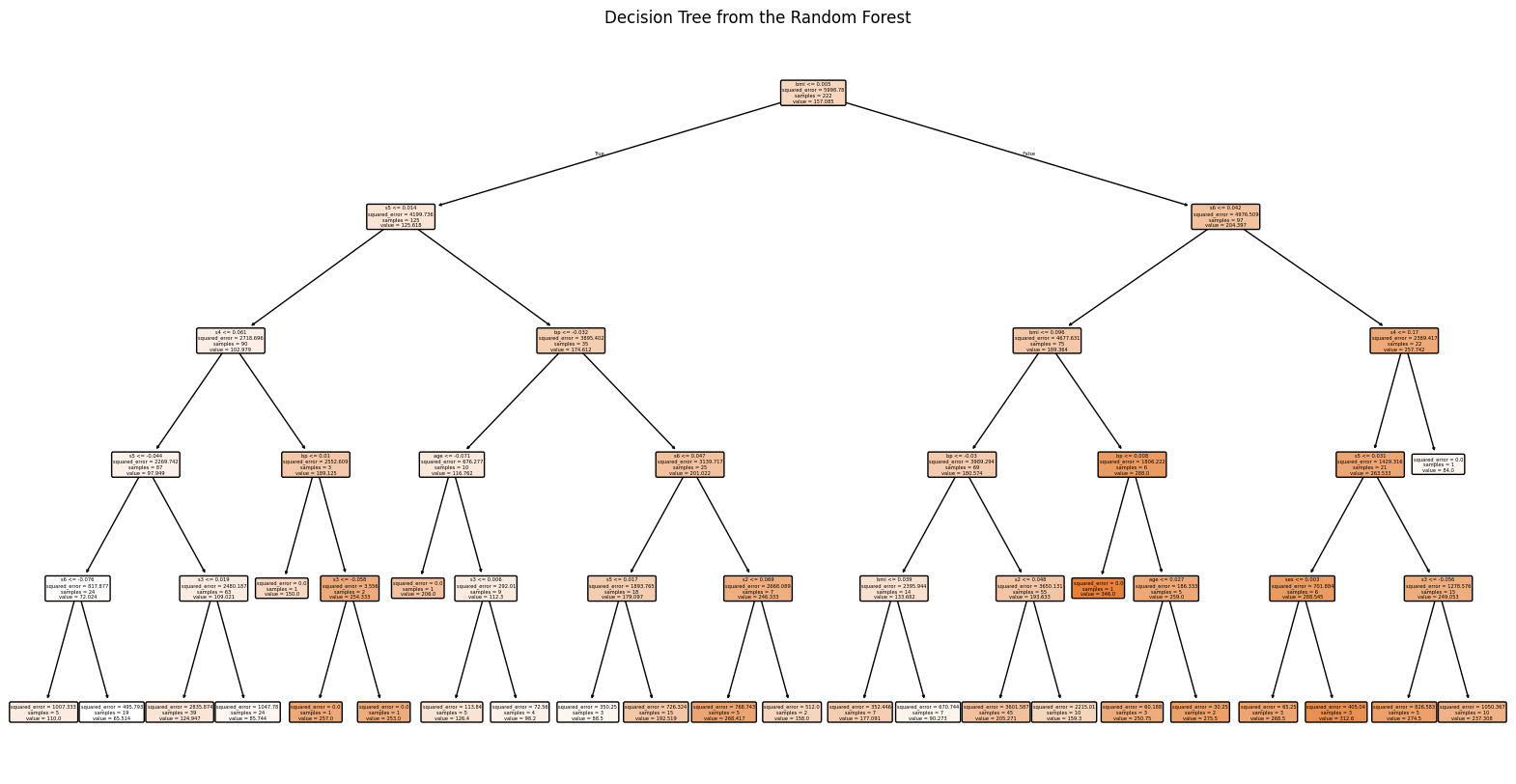
Plot with graphviz
tree = rf_regressor.estimators_[0] # Define feature names (the diabetes dataset does not provide feature names directly) features = diabetes.feature_names # Export the selected tree to DOT format dot_file_path = "diabetes_forest_tree.dot" export_graphviz( tree, out_file=dot_file_path, feature_names=features, # Feature names from the diabetes dataset filled=True, # Color the nodes rounded=True, # Rounded corners for nodes special_characters=True # Allows special characters in labels ) # Read the DOT file and visualize the tree using Graphviz with open(dot_file_path) as f: dot_graph = f.read() # Visualize the tree using graphviz graph = graphviz.Source(dot_graph) graph.render("diabetes_forest_tree") # Save the tree visualization as a file graph
Plot with dtreeviz
from dtreeviz import model tree = rf_regressor.estimators_[0] viz_model = model( tree, X_train=X_train, y_train=y_train, feature_names=diabetes.feature_names, target_name="diabetes", ) viz_model.view()
Plot with supertree
In previous example tree is a little unreadable. supertree has simmilar problem, but fixes it. I can use scroll in my mouse to zoom in on particular elements and see details on interactive diagrams.
You can try this out on mljar.com/supertree.
from supertree import SuperTree st = SuperTree( rf_regressor, X_train, y_train, diabetes.feature_names, "diabetes" ) st.show_tree(which_tree=0)
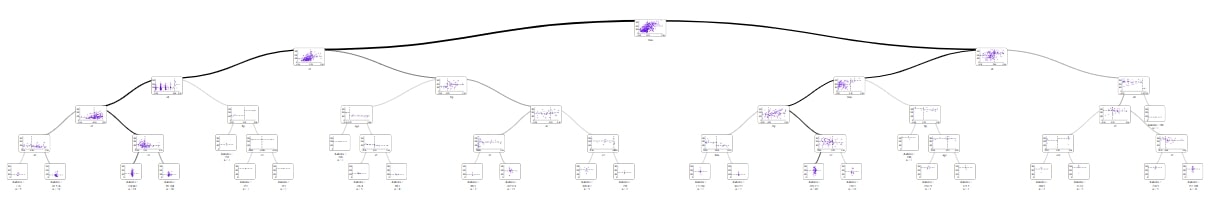
supertree package looks like great all-rounder, especially in notebooks. It not only looks interesting but also gives easy acces to details by hovering mouse over elements. However I think that this interactivity is what rises supertree over other comeptitors.
I can not recommed this package enough.
Run AI for Machine Learning — Fully Local
MLJAR Studio is a private AI Python notebook for data analysis and machine learning. Generate code with AI, run experiments locally, and keep full control over your workflow.
Related Articles
- Create Retrieval-Augmented Generation using OpenAI API
- Transfer data from Postgresql to Google Sheets
- Machine Learning Algorithm Comparison
- TabNet vs XGBoost
- 4 Effective Ways to Visualize LightGBM Trees
- 4 Effective Ways to Visualize XGBoost Trees
- Is weather correlated with cryptocurrency price?
- How to become a Data Scientist?
- 6 best packages for data visualization in Python
- How to install packages in Python